About_
I'm a DPhil (PhD) student in the Department of Statistics,
broadly interested in machine learning approaches to biomolecular structure and design.
My research in the Oxford Protein Informatics Group (OPIG) focusses on advancing de novo antibody design using generative models.
In this work, I am fortunate to be supervised by Prof Charlotte Deane,
and Matthew Raybould,
as well as Rebecca Croasdale-Wood at AstraZeneca.
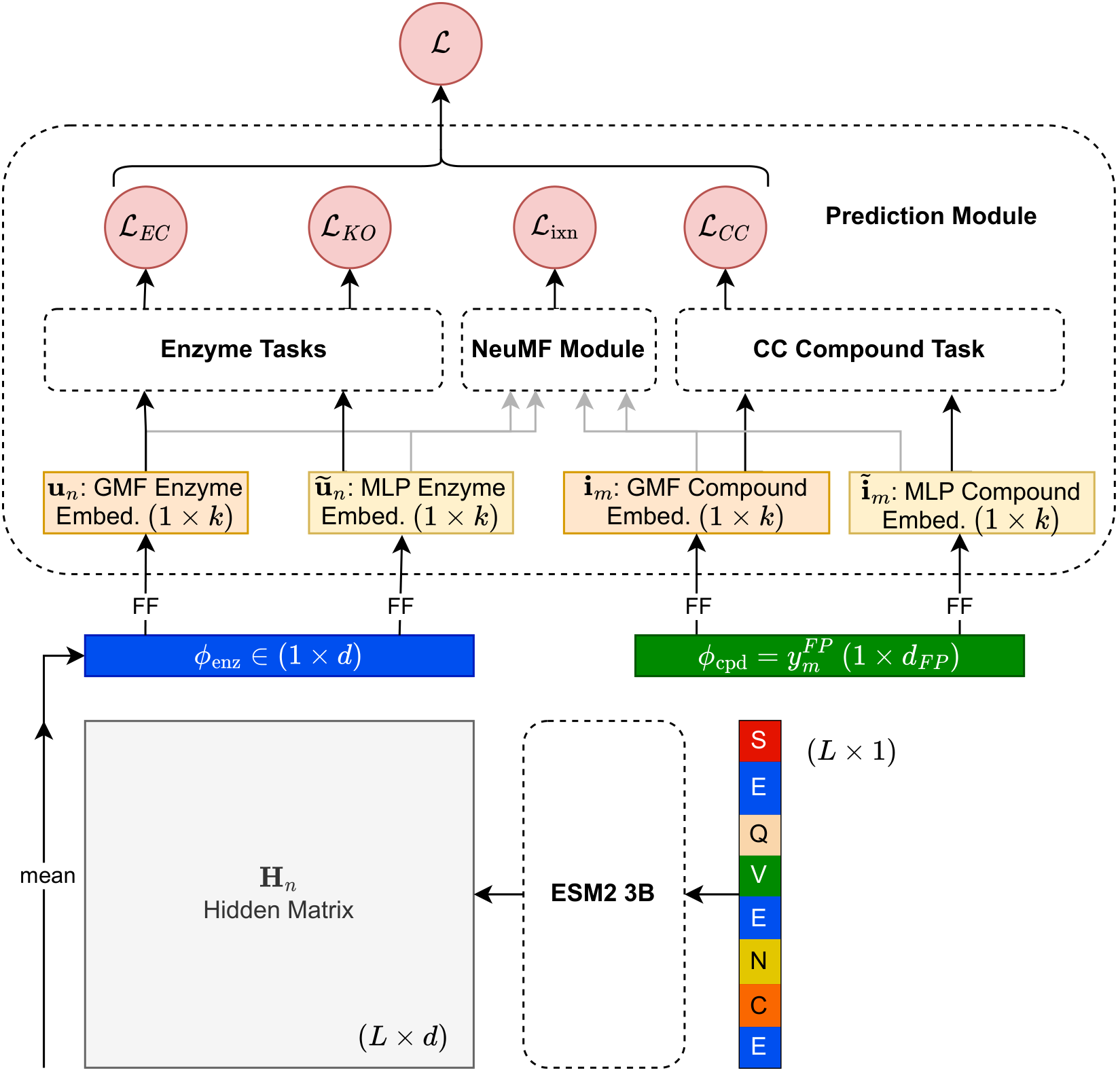
In the past, I've tackled enzyme-substrate interaction prediction using protein language models, and also had a brief stint in theoretical chemical physics,
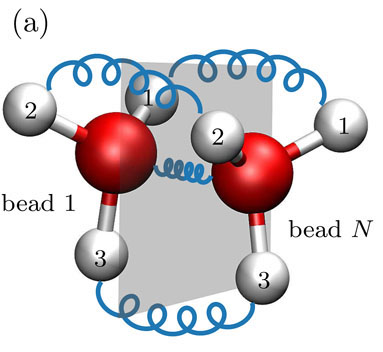
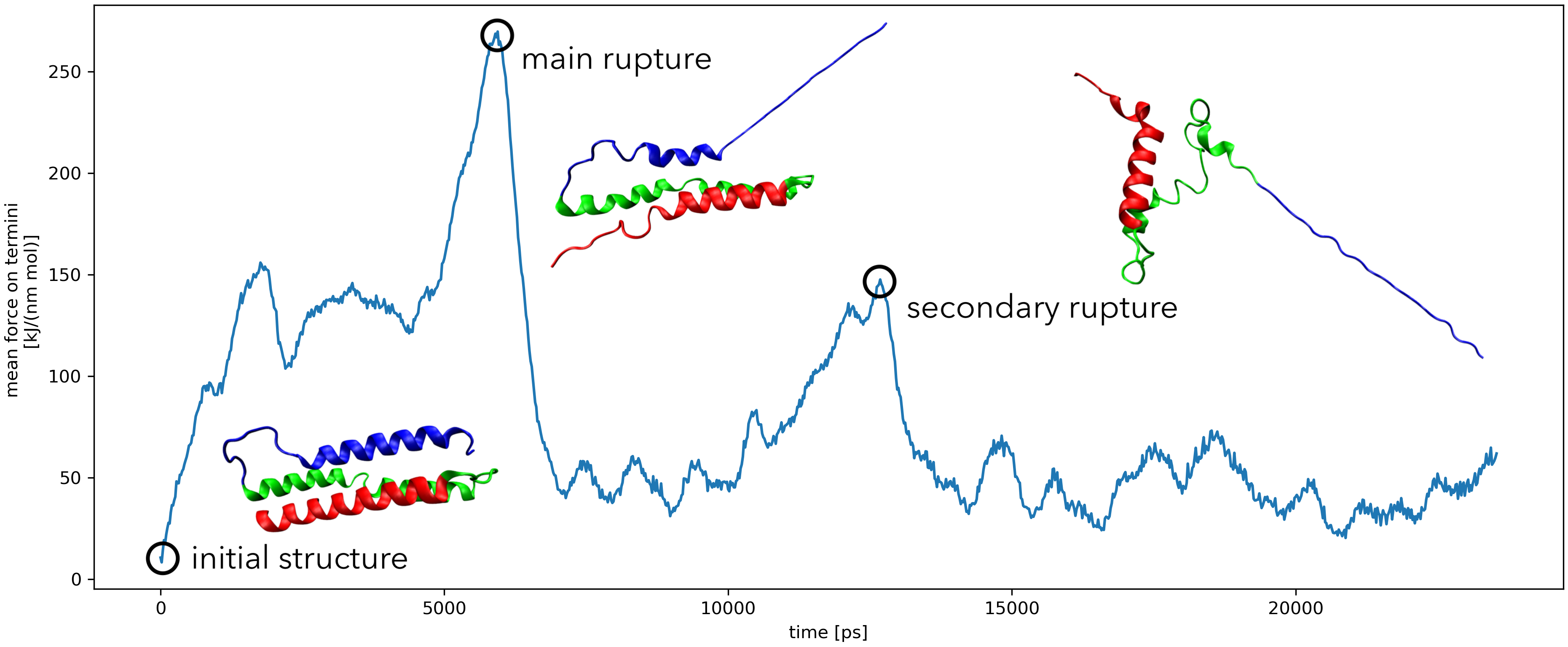
predicting quantum tunnelling splittings using classical molecular dynamics.
Check out the research and publications pages for more details.
As a hobby, I've occasionally dabbled in information-theoretic approaches to evolution, and game theory on graphs.
My DPhil work is generously funded by Oxford University's flagship
Clarendon Scholarship, alongside additional partnership awards by Oxford University, Balliol College, and AstraZeneca.
Throughout my BSc and MSc, I had the privilege to receive support from the
German Academic Scholarship Foundation.
Education_

2023-2027
DPhil at the Department of Statistics (SABS:R3 CDT)
Balliol College, University of Oxford

2020-2023
MSc Computational Biology & Bioinformatics
ETH Zürich, Switzerland

2015-2018
BSc Biochemistry
Heidelberg University, Germany